Deep neural network models form the backbone of most state-of-the-art image analysis and natural language processing algorithms. With the recent development of large-scale deep learning techniques such as data and model parallelism, large convolutional neural network (CNN) models can be trained on datasets of millions of images in minutes. However, applying a CNN model on ultra-high resolution images, such as 3D computed tomography (CT) images that can have up to 108 pixels, remains challenging. With existing techniques, a processor still needs to host a minimum of 32GB of partial, intermediate data, whereas individual GPUs or TPUs typically have only 12-32GB memory. A typical solution is to process image patches separately from one another, which leads to complicated implementation and sub-optimal performance due to information loss.
In “High Resolution Medical Image Analysis with Spatial Partitioning”, a collaboration with the Mayo Clinic, we push the boundary of massive data and model parallelism through use of the Mesh-TensorFlow framework, and demonstrate how this technique can be used for ultra-high resolution image analysis without compromising input resolution for practical feasibility. We implement a halo exchange algorithm to handle convolutional operations across spatial partitions in order to preserve relationships between neighboring partitions. As a result, we are able to train a 3D U-Net on ultra-high resolution images (3D images with 512 pixels in each dimension), with 256-way model parallelism. We have additionally open-sourced our Mesh-TensorFlow-based framework for both GPUs and TPUs for use by the broader research community.
Data and Model Parallelism with Mesh-TensorFlow
Our implementation is based on the Mesh-TensorFlow framework for easy and efficient data and model parallelism, which enables users to split tensors across a mesh of devices according to the user defined image layout. For example, users may provide the mesh of computational devices as 16 rows by 16 columns for a total of 256 processors, with two cores per processor. They then define the layout to map the spatial dimension x of their image to processor rows, map spatial dimension y to processor columns, and map the batch dimension (i.e., the number of image segments to be processed simultaneously) to cores. The partitioning and distributing of a training batch is implemented by Mesh-TensorFlow at the tensor level, without users worrying about implementation details. The figure below shows the concept with a simplified example:
Spatial Partitioning with Halo Exchange
A convolution operation executed on an image often applies a filter that extends beyond the edge of the frame. While there are ways to address this when dealing with a single image, standard approaches do not take into account that for segmented images information beyond the frame edge may still be relevant. In order to yield accurate results, convolution operations on an image that has been spatially partitioned and redistributed across processors must take into account each image segment’s neighbors.
One potential solution might be to include overlapping regions in each spatial partition. However, since there are very likely many subsequent convolutional layers and each of them introduces overlap, the overlap will be relatively large — in fact, in most cases, the overlap could cover the entire image. Moreover, all overlapping regions must be included from the start, at the very first layer, which may run into the memory constraints that we are trying to resolve.
Our solution is totally different: we implemented a data communication step called halo exchange. Before every convolution operation, each spatial partition exchanges (receives and sends) margins with its neighbors, effectively expanding the image segment at its margins. The convolution operations are then applied locally on each device. This ensures that the result of the convolutions for the whole of the image remain identical with or without spatial partitioning.
Proof of Concept – Segmentation of Liver Tumor CT Scans
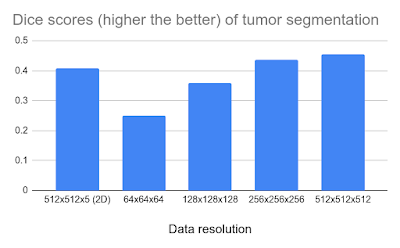
We then applied this framework to the task of segmenting 3D CT scans of liver tumors (LiTS benchmark). For the evaluation metric, we use the Sørensen–Dice coefficient, which ranges from 0.0 to 1.0 with a score of 0 indicating no overlap between segmented and ground truth tumor regions and 1 indicating a perfect match. The results shown below demonstrate that higher data resolution yields better results. Although the return tends to diminish when using the full 5123 resolution (512 pixels in each of x, y, z directions), this work does open the possibility for ultra-high resolution image analysis.
Conclusion
Existing data and model parallelism techniques enabled the training of neural networks with billions of parameters, but cannot handle input images above ~108 pixels. In this work, we explore the applicability of CNNs on these ultra-high resolution images, and demonstrate promising results. Our Mesh-TensorFlow-based implementation works on both GPUs and TPUs, and with the released code, we hope to provide a possible solution for some previously impossible tasks.
This article has been published from the source link without modifications to the text. Only the headline has been changed.
Source link




