In order to predict chemical reactions, researchers have created a platform that combines AI and automated experimentation. This could expedite the process of designing novel drugs.
For the creation of new medications, it is essential to predict how molecules will react; but, in the past, this has been a trial-and-error procedure, and the reactions frequently fail. Chemists typically use computationally expensive and frequently erroneous simplified models of electrons and atoms to anticipate how molecules will react.
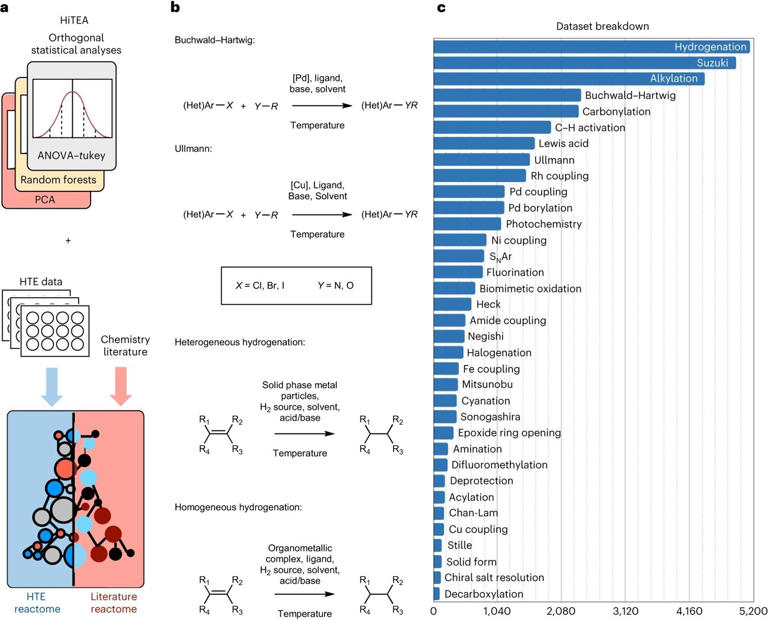
In order to comprehend chemical reactivity, researchers at the University of Cambridge have now created a data-driven method that is motivated by genomics and combines machine learning with automated experimentation, significantly accelerating the process. Their method has been dubbed the “chemical reactome,” as it was verified using a dataset including over 39,000 reactions that are relevant to the pharmaceutical industry.
Their findings, which were published in the journal Nature Chemistry, are the outcome of Cambridge and Pfizer’s cooperation.
The reactome has the potential to revolutionize our understanding of organic chemistry, according to the paper’s first author, Dr. Emma King-Smith of Cambridge’s Cavendish Laboratory. We could produce medications and a plethora of other beneficial goods far more quickly if we had a greater understanding of the chemistry. More importantly, though, anyone who works with molecules will benefit from the knowledge we want to produce.
The reactome method identifies pertinent relationships between reactants, reagents, and reaction performance from the data and highlights areas where the data is lacking. The data comes from automated trials that run quickly, or at a high throughput.
The reactome method identifies pertinent relationships between reactants, reagents, and reaction performance from the data and highlights areas where the data is lacking. The data comes from automated trials that run quickly, or at a high throughput.
High throughput chemistry has changed the game, but according to King-Smith, They thought there was a way to learn more about chemical reactions than what can be inferred from the preliminary findings of a high throughput experiment.
According to Dr. Alpha Lee, the research’s principal investigator, their approach reveals the hidden relationships between reaction components and outcomes. The enormous dataset that they used to train the model will contribute to advancing chemical discovery from the era of trial and error to the era of big data.
In a separate work that was published in Nature Communications, the scientists created a machine learning method that allows chemists to quickly construct drugs by introducing exact modifications to pre-specified molecular areas.
This method spares chemists from having to start from scratch when adjusting complex molecules, such as a last-minute design modification. In the lab, creating a molecule usually involves several steps, much like constructing a house. The standard procedure for chemists to change a molecule’s core is to reconstruct it, much like when they rebuild a house from the ground up. Core differences are crucial for the creation of medicines, nevertheless.
Late-stage functionalization reactions are a family of reactions that aim to avoid starting from scratch by introducing chemical alterations directly to the core. Making late-stage functionalization selective and regulated is tough, though, as there are usually a lot of molecular areas that can respond, and the results are unpredictable.
Late-stage functionalizations can have unexpected outcomes, and existing modeling techniques—including our own professional intuition—aren’t flawless, according to King-Smith. Better screening would be possible with a more predictive model.
A machine learning model that the researchers created forecasts the location of a molecule’s reaction as well as how the reaction site changes in response to various reaction conditions. Chemists can now accurately modify a molecule’s core thanks to this.
Before fine-tuning the model to predict these complex changes, King-Smith explained that we pretrained it on a sizable amount of spectroscopic data, essentially teaching it general chemistry. Because there aren’t many reports of late-stage functionalization reactions in the scientific literature, the team was able to circumvent the data limitation with this strategy. Through experimental validation on a wide range of drug-like compounds, the team was able to predict the locations of reactivity with high accuracy across a range of situations.
According to Lee, the issue of little data in relation to the vastness of chemical space frequently hinders the use of machine learning to chemistry. Their methodology—building models that learn from sizable datasets that are comparable to but distinct from the issue they are attempting to solve—solves this basic low-data obstacle and may pave the way for developments beyond late-stage functionalization.








