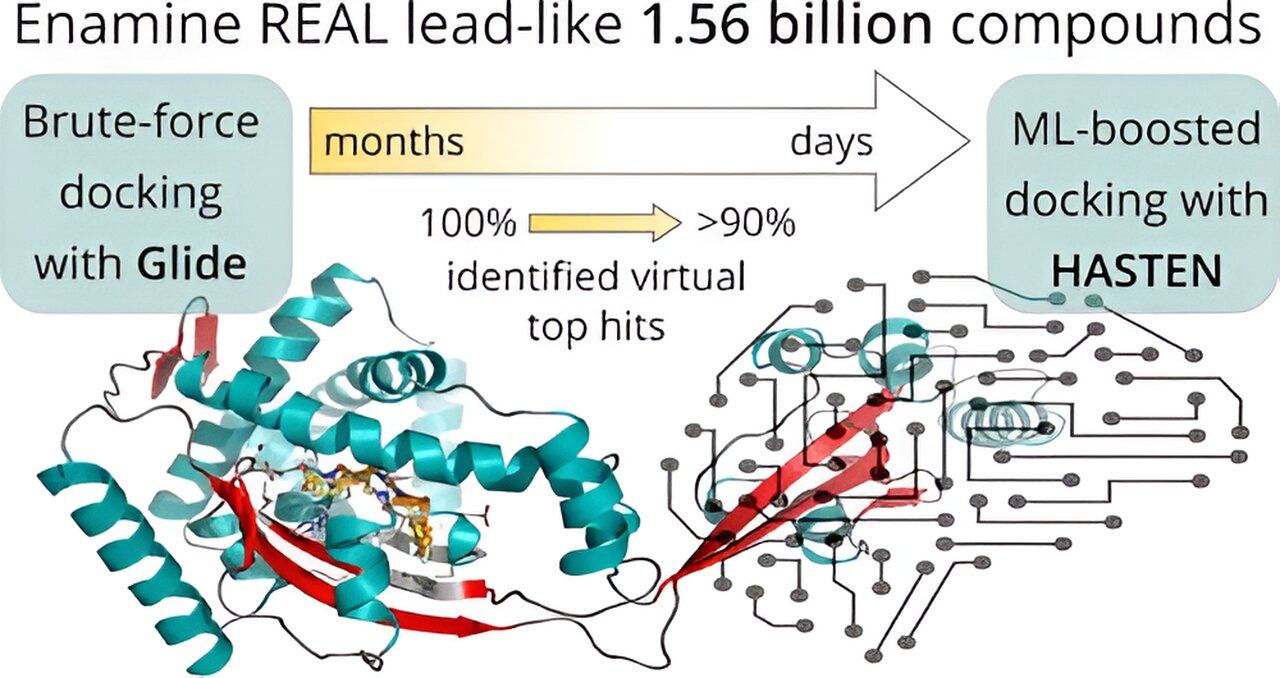
The processing of 1.56 billion drug-like molecules was accelerated by machine learning, which resulted in a 10-fold time reduction. One of the largest virtual drug screens ever conducted was by University of Eastern Finland researchers in collaboration with companies and supercomputers.
Researchers frequently use quick computer-assisted screening of huge chemical libraries to locate compounds that can block a therapeutic target in their search for novel pharmacological molecules. An enzyme that enables a bacterium to resist antibiotics or a virus to infect its host are two examples of such a target. Over the past few years, there has been a dramatic increase in the size of these clusters of tiny organic molecules.
Even with the use of cutting-edge supercomputers, the screening of a modern billion-scale compound library against just one drug target can take many months or years due to libraries expanding faster than the speed of the machines needed to process them. Therefore, it is obvious that speedier methods are essential.
In a study that was published in the Journal of Chemical Information and Modelling, Dr. Ina Pöhner and colleagues from the School of Pharmacy at the University of Eastern Finland joined forces with CSC—IT Centre for Science Ltd., the organization that hosts Finland’s powerful supercomputers, and commercial partners from Orion Pharma to investigate the potential of machine learning in the acceleration of giga-scale virtual screens.
The researchers first created a baseline: Before using artificial intelligence to speed up the screening, they did the following: With the aid of the supercomputers Mahti and Puhti, as well as molecular docking, 1.56 billion drug-like compounds were assessed against two pharmacologically important targets over the course of roughly six months in an unparalleled virtual screening effort. Docking is a computer method that places the tiny molecules into the target’s binding area and determines a “docking score” to indicate how well they fit. So, all 1.56 billion molecules’ docking scores were initially calculated.
The outcomes were then contrasted with a machine learning-boosted screen utilizing HASTEN, a technology created by co-author of the paper and Orion Pharma’s Dr. Tuomo Kalliokoski.
Machine learning is used by HASTEN to understand the characteristics of molecules and how those characteristics affect how well the compounds score. The machine learning model can predict docking scores for additional compounds in the library considerably more quickly than the brute-force docking strategy when given enough samples from conventional docking, according to Kalliokoski.
In fact, the technology successfully identified 90% of the highest-scoring compounds in less than 10 days with just 1% of the entire library docked and utilized as training data.
The study was the first comprehensive giga-scale comparison of a machine learning-boosted docking tool with a traditional docking baseline. The majority of the top-scoring compounds identified by conventional docking could be reliably and frequently reproduced by the machine learning-boosted technique, according to Pöhner, in a noticeably shorter amount of time.
This initiative is a great illustration of how academics and industry can work together and how CSC can provide one of the best computing resources available. We were able to accomplish our lofty objectives by combining our knowledge, expertise, and technology, according to Professor Antti Poso, who directs the computational drug discovery team at the University of Eastern Finland’s DrugTech Research Community.
Comparable studies are still difficult to find in most contexts. As a result, the authors made substantial datasets produced for the study available to the general public. With 1.56 billion compound-docking results for two targets that can be utilized as benchmarking data, their ready-to-use screening library for docking enables others to accelerate their individual screening efforts.
This information will promote the creation of time and resource-saving methods in the future, advancing the field of computational drug discovery.