Help the global community to better understand the disease by mining mountain sized body of research.
The Project
In response to the COVID-19 pandemic, the White House and a coalition of leading research groups have prepared the COVID-19 Open Research Dataset.
This is a collection of large number of research papers on COVID-19. The hope is that the machine learning community can help extract useful insights from this large collection of research papers.
The data and detailed description can be found here a Kaggle.
What Is The Idea?
The idea is to create a language model trained on the “body text” of the research paper available in this dataset. The resulting language model can then be used to build a classifier of some sort.
Currently we don’t have any labelled dataset of COVID-19 data but I hope that in case a labelled dataset is available then we can use this language model to do some sort of classification task.
Even if a labelled dataset is not available we can still customize this language model (or an improved version of this) to predict the next sequence of words for a search term and gain hidden insights which would otherwise hide from the human eye.
Importing the necessary libraries
I am using fastai to create the language model. So fast.text is imported. Json is used to parse json data.
from fastai.text import *
import numpy as np
import json
Let’s Set All The Paths?
Let’s set all the path which is required to work on our data.
pathis the root input directory.outputPathis where we would be saving our csv , models etc.datasetPathis the directory to the dataset.
path = Path('/kaggle/input')
outputPath = Path('/kaggle/working')
datasetPath = path/'CORD-19-research-challenge/'
Let’s also set the path to the research paper collection. For this language model we will select a subset of the entire corpus. This will help us in quick initial testing of our code.
pmcCustomLicenseis the path to the directory which contains the json files for the research papers sourced from pmc.biorxivMedrxivis the path to the directory which contains the json files for the research papers sourced from biorxiv-Medrxiv.
pmcCustomLicense = datasetPath/'custom_license/custom_license'
biorxivMedrxiv = datasetPath/'biorxiv_medrxiv/biorxiv_medrxiv'
How To Parse The Data From Json?
The data from the research papers are stored as json in this dataset. We need to parse the data from the json files before we use that.
For this project I am trying to build a language model and currently I am interested in the “body_text” key of the json files.
This is where the body of the research papers are present.
Here I have made used of the functions and techniques already used in this notebook. I am grateful to the creator of this notebook for this.
I have made a few customization to the functions from the original notebook but almost all of it remains the same as the original content.
Here the Idea is to parse the json files to extract bodytext into a pandas dataframe.
The functions do the following →
load_files() –
- Takes a tuple as an argument. This tuple is a collection of lists having the path the directories from where the json needs to be fetched.
- Then this function goes through each of these directories and appends them to a new list fileList. this way the contents of the original lists (i.e. the filpaths) becomes a part of a bigger list fileList.
example usage
filePathList1 = [‘path1’, ‘path2’, ‘path3’] filePathList2 = [‘path4’, ‘path5’, ‘path6’]load_files(filePathList1, filePathList2)Output → [‘path1’, ‘path2’, ‘path3’, ‘path4’, ‘path5’, ‘path6’]def load_files(fileNames: tuple): fileList = []for file in fileNames: fileList.append(file) return fileList
Then we call the function with the preferred directory where the research papers are stored.
files = load_files((biorxivMedrxiv.iterdir()))
You can notice that I didn’t pass the biorxivMedrxiv as is. The reason is because biorxivMedrxiv is a “posixpath” object and this is not an iterable object. To iterate through the biorxivMedrxiv I used the iterdir() method which gives us an iterator.
The file paths processed with the load_files() contains path objects and may not be suitable for certain operations.
For example path objects can’t be iterated over as the contents returned by load_files() are “posixpath” objects.
We will convert those path objects into their string “paths” with the filePath() function. This function does the following →
- Takes in the list having the path objects.
- Iterates through the list and converts each list item i.e. the path objects into string format.
- Then it puts those string paths into the filePath list.
Note: For the load_files() function I have passed in the file list tuple as an iterdir() so that we can iterate over these paths. So, I think that the contents returned by the load_files() function can be iterated over without the need of the filePath() function. However I have’t tested this yet.
def filePath(files):
filePath = []
for file in files:
filePath.append(str(file))
return filePath
Then we call the function.
filePaths=filePath(files)
getRawFiles() function –>
- Takes in a list of file paths
- Goes through each file path from the list
filesand uses thejson.load()method to read the contents. - finally it appends these json object into the
rawFileslist.
def getRawFiles(files: list):
rawFiles = []
for fileName in files:
rawFile = json.load(open(fileName, 'rb'))
rawFiles.append(rawFile)
return rawFiles
Calling this function would give us the raw contents of the json files.
rawFiles = getRawFiles(filePaths)
format_name() →
- Takes in the json object with author as the key.
- Joins the names in the following sequence <first name> <middle name> <last name.
- Function also takes care o fthe fact that if there no middle name then just do <first name> <last name
format_affiliation() →
- Takes in the json object with key as affiliation.
- If location details are there in json then put it into a list and return it.
- If institution is there then join location and instituion details together and put it into a list.
format_authors()→
- Takes in the json object with key as author.
- Joins the author’s name with the affiliations if affiliations are available.
format_body() →
- Takes in the json object with key as body_text.
- Extracts the text and then appends it into a list.
format_bib() →
- Takes in the json object with key as bib.
- Joins the ‘title’, ‘authors’, ‘venue’, ‘year’ together to form a string.
def format_name(author): middle_name = " ".join(author['middle']) if author['middle']: return " ".join([author['first'], middle_name, author['last']]) else: return " ".join([author['first'], author['last']])def format_affiliation(affiliation): text = [] location = affiliation.get('location') if location: text.extend(list(affiliation['location'].values())) institution = affiliation.get('institution') if institution: text = [institution] + text return ", ".join(text)def format_authors(authors, with_affiliation=False): name_ls = [] for author in authors: name = format_name(author) if with_affiliation: affiliation = format_affiliation(author['affiliation']) if affiliation: name_ls.append(f"{name} ({affiliation})") else: name_ls.append(name) else: name_ls.append(name) return ", ".join(name_ls)def format_body(body_text): texts = [(di['section'], di['text']) for di in body_text] texts_di = {di['section']: "" for di in body_text} for section, text in texts: texts_di[section] += textbody = ""for section, text in texts_di.items(): body += section body += "\n\n" body += text body += "\n\n" return bodydef format_bib(bibs): if type(bibs) == dict: bibs = list(bibs.values()) bibs = deepcopy(bibs) formatted = [] for bib in bibs: bib['authors'] = format_authors( bib['authors'], with_affiliation=False ) formatted_ls = [str(bib[k]) for k in ['title', 'authors', 'venue', 'year']] formatted.append(", ".join(formatted_ls))return "; ".join(formatted)
generate_clean_df() –>
- Uses the above helper functions to create a pandas dataframe.
- The resulting dataframe would have the following columns -> ’paper_id’, ‘title’, ‘authors’, ‘affiliations’, ‘abstract’, ‘text’, ‘bibliography’, ‘raw_authors’, ‘raw_bibliography’
def generate_clean_df(all_files): cleaned_files = [] for file in all_files: features = [ file['paper_id'], file['metadata']['title'], format_authors(file['metadata']['authors']), format_authors(file['metadata']['authors'], with_affiliation=True), format_body(file['abstract']), format_body(file['body_text']), format_bib(file['bib_entries']), file['metadata']['authors'], file['bib_entries'] ]cleaned_files.append(features)col_names = ['paper_id', 'title', 'authors', 'affiliations', 'abstract', 'text', 'bibliography','raw_authors','raw_bibliography']clean_df = pd.DataFrame(cleaned_files, columns=col_names) clean_df.head() return clean_df
Then calling this function would generate our dataframe.
pd.set_option('display.max_columns', None)
cleanDf = generate_clean_df(rawFiles)
cleanDf.head()
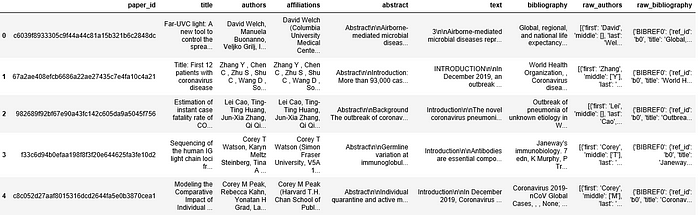
Finally save the dataframe to a csv file. Since the entire previous process takes a bit of time, so saving the file to csv saves time later.
cleanDf.to_csv(outputPath/'cleandf.csv')
The Data
We then create a databunch from the csv that we created from the json files.
The createDataBunchForLanguageModel() is a helper function. This does the following →
- Creates a databunch from the csv file.
- The data in the databunch is fetched from the ‘text’ column of the csv file.
- 10% of the data is reserved as the validation data.
- Since the language model that we will create down the line will be a self-supervised model. It takes in the label from the data itself.
- Thus the label_for_lm() helps us with that.
- After the databunch is created, it is saved to a pickle file.
- Next time when this code runs we don’t need to go through the process of creation of the databunch once again. We can use the pickle file.
To know more about “databunch” refer to my earlier article here.
def createDataBunchForLanguageModel(outputPath: Path,
csvName: str,
textCol: str,
pickelFileName: str,
splitBy: float,
batchSize: int):
data_lm = TextList.from_csv(outputPath,
f'{csvName}.csv',
cols=textCol)\
.split_by_rand_pct(splitBy)\
.label_for_lm()\
.databunch(bs=batchSize)
data_lm.save(f'{pickelFileName}.pkl')
Calling this function to create the databunch.
createDataBunchForLanguageModel(outputPath,
'cleandf',
'text',
'cleanDf',
0.1,
48)
The loadData() is a helper function to load the databunch file. It takes in the required batch size which we want to load from the databunch.
def loadData(outputPath: Path,
databunchFileName: str,
batchSize: int,
showBatch: bool= False):
data_lm = load_data(outputPath,
f'{databunchFileName}.pkl',
bs=batchSize)
if showBatch:
data_lm.show_batch()
return data_lm
We will use this function shortly.
Building The Learner
We use ULMFIT to create a language model on the research data corpus and then fine tune this language model.
We create the language model learner. The language_model_learner() is a fastai method which helps us to build a learner object with the data created in the previous sections.
Here we use a batch size of 48. This learner is built using a pretrained language model architecure AWD_LSTM. This is the self supervised part. This is a model which was trained on an English language corpus to predict the next sequence of sentences and thus understands the structure of the language.
We just need to fine tune this model on our data corpus which then can be used to build a classifier.
I am not building classifier yet because I don’t have any idea as to what we need to classify. Yet I believe that this fine tuned language model can be customized to find hidden information in the large corpus of research papers which would otherwise be hidden/missed by the human readers.
Note: I am not diving deep into ULMFIT as that is not the aim of this post. I may do a detailed post on it later. However a detailed explanation of ULMFIT can be found here at fastai.
learner = language_model_learner(loadData(outputPath,
'cleanDf',
48,
showBatch= False),
AWD_LSTM,
drop_mult=0.3)
We plot the learning rate for our language model. From this plot we will try to find the best learning rate suitable for our model. The plotLearningRate() is a helper function which plots the learning rate for us.
An explanation of how to use the learning rate can be found here.
def plotLearningRate(learner, skip_end):
learner.lr_find()
learner.recorder.plot(skip_end=skip_end)
We plot the learning rate now.
plotLearningRate(learner, 15)
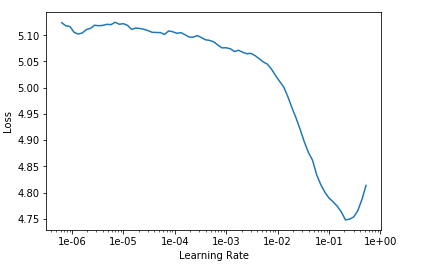
We then take in that learning rate as our start where the plot diverges. The model is then trained with the “fit one cycle” policy with this starting learning rate.
This is where we train only the head.
learner.fit_one_cycle(1, 1e-02, moms=(0.8,0.7))
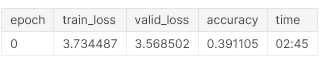
Since we are creating a language model, we are not overly concerned about getting the best possible accuracy here. We unfreeze the network and train some more.
learner.unfreeze()
learner.fit_one_cycle(10,1e-3, moms=(0.8,0.7))
Finally we save the fine tuned language model for later use in testing/prediction.
learner.save('fineTuned')
Testing
Let’s see if the model can connect the information/knowledge from the corpus. We first load the saved model and then try to find prediction for a search term.
learner = learner.load('fineTuned')
Let’s take a test sentence as our input.
TEXT = "Range of incubation periods for the disease in humans"
N_WORDS = 40
N_SENTENCES = 2
We will see if our model can predict related words to our test string.
print("\n".join(learner.predict(TEXT, N_WORDS, temperature=0.75) for _ in range(N_SENTENCES)))

We have a pretty good language model available now. Now, that we have fine tuned our language model , we can save the model which predicts the next sequence of words and the encoder which is responsible for creating and updating the hidden states.
The saved model can be used for transfer learning in further tasks and the saved encoder can be used to build a classifier fo rtext classification on any COVID-19 related labelled dataset.
We will use .save() to save the .pth file.
The save() function mentioned below is a helper function which can save the model only or the encoder and model in separate files.
def save(learner,
saveEncoder: bool = True):
if saveEncoder:
learner.save_encoder('fine_tuned_encoder')
learner.save('fine_tuned_model')
calling this function will do the saving for you.
save(learner)
End Notes
I hope that this kernel and this language model would be helpful for someone else who might be working on this dataset or any COVID-19 related data.
I am hoping that someone who is more seasoned in NLP and deep learning would improve on this model and bring out something useful which could then contribute towards fighting COVID-19
I am in no way an expert in NLP so the network that I have developed here is based on the code from the lesson3 of the course — “Practicel Deep Learning For coders” by fastai.
I am hoping that someone who is more seasoned in NLP and deep learning would improve on this model and bring out something useful which could then contribute towards giving us a better chance in fighting COVID-19.
The code used in this post is available as a kaggle kernel here and was submitted as part of one of the task challenges for the COVID-19 research data.




